4.12. Chapter 4 Spectroscopy#
4.13. Generation of Characteristic X-Rays#
part of
MSE672: Introduction to Transmission Electron Microscopy
Spring 2024
| Gerd Duscher | Khalid Hattar |
| Microscopy Facilities | Tennessee Ion Beam Materials Laboratory |
| Materials Science & Engineering | Nuclear Engineering |
| Institute of Advanced Materials & Manufacturing | |
Background and methods to analysis and quantification of data acquired with transmission electron microscopes.
4.13.1. Load relevant python packages#
4.13.1.1. Check Installed Packages#
import sys
import importlib.metadata
def test_package(package_name):
"""Test if package exists and returns version or -1"""
try:
version = importlib.metadata.version(package_name)
except importlib.metadata.PackageNotFoundError:
version = '-1'
return version
if test_package('pyTEMlib') < '0.2024.2.3':
print('installing pyTEMlib')
!{sys.executable} -m pip install --upgrade pyTEMlib -q
print('done')
done
4.13.2. First we import the essential libraries#
All we need here should come with the annaconda or any other package
%matplotlib widget
import matplotlib.pyplot as plt
import numpy as np
import sys
import os
if 'google.colab' in sys.modules:
from google.colab import output
from google.colab import drive
output.enable_custom_widget_manager()
from pyTEMlib.xrpa_x_sections import x_sections
__notebook__ = 'CH4_14-Characteristic-X_Rays'
__notebook_version__ = '2019_08_21'
4.13.3. X-Ray Families#
The case is not always as simple as in the case of the carbon atom in the Introduction
As the atomic number
For example a Na atom which has an M-shell The 1s state can be filled by an L- or an M-shell, producing two different characteristic X-rays:
K-L
K-M
For heavier atoms than sodium more options exist:
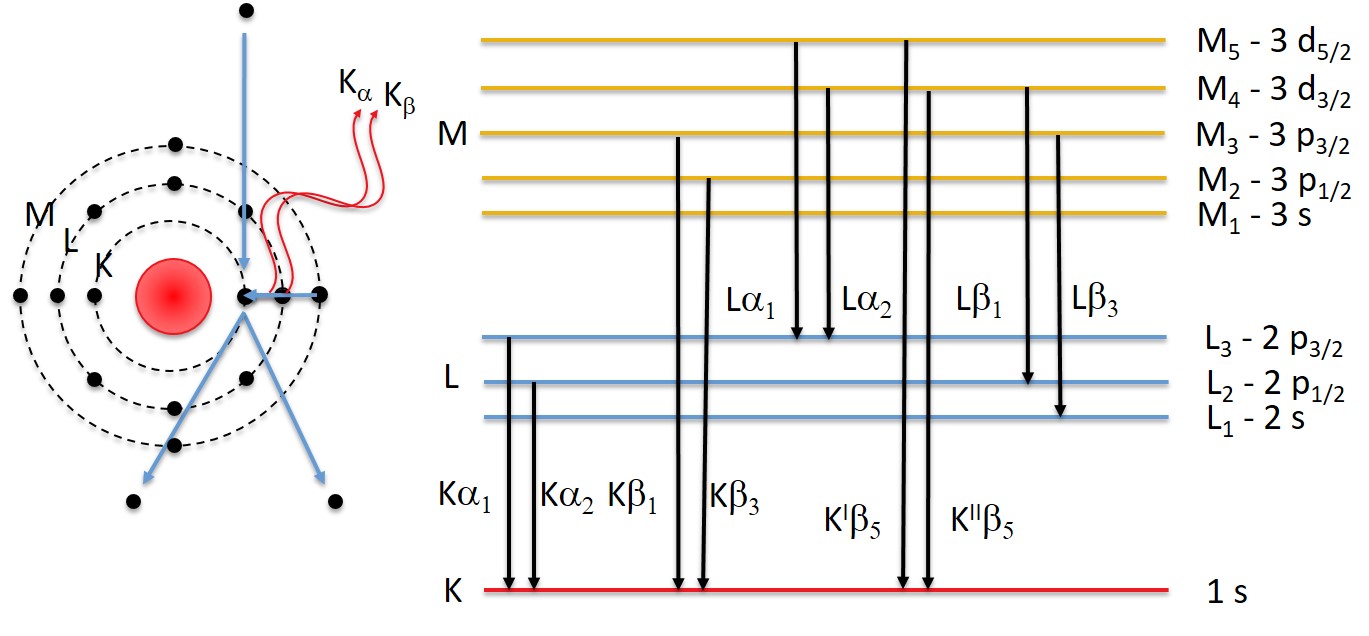
4.13.4. X-ray Nomenclature#
Two systems are in use for naming characteristic X-ray lines.
The oldest one is the Siegbahn system in wich first the shell where the ionization occured is listed and then a greek letter that suggests the order of their families by their relative intensity (
For closely related memebers of a family additionally numebrs are attched such as
While most commercial software still uses the Siegbahn system the International Union of Pure and Applied Chemistry (IUPAC) has officially changed the system to a protocol that first dentoes the subshell were the ionization occured and followed with the term that indicates from which subshell the relaxation originates.
So
Siegbahn |
—–IUPAC—– |
Siegbahn |
—–IUPAC—– |
Siegbahn |
—–IUPAC—– |
|---|---|---|---|---|---|
4.13.5. X-ray Weight of Lines#
Within the excitation probaility of the characteristic X-ray lines are not equal.
For sodium (see above for energies),
the ratio of
This ratios between the lines are a strong function of the atomic number
We load the data of the X-Ray lines from data from NIST, the line weights are taken from the Program DTSA-II, which is the quasi-standard in EDS analysis.
K_M5 = []
ele_K_M5 = []
L3_M1 = []
ele_M3_M1 = []
L1_N3 = []
ele_L1_N3 = []
L3_N5 =[]
ele_L3_N5 = []
L2_M4 = []
ele_L2_M4 = []
for element in range(5,82):
if 'K-M5' in x_sections[str(element)]['lines']:
K_M5.append(x_sections[str(element)]['lines']['K-M5']['weight']/x_sections[str(element)]['lines']['K-L3']['weight'])
ele_K_M5.append(element)
if 'L3-M5' in x_sections[str(element)]['lines']:
if 'L3-M1' in x_sections[str(element)]['lines']:
L3_M1.append(x_sections[str(element)]['lines']['L3-M1']['weight']/x_sections[str(element)]['lines']['L3-M5']['weight'])
ele_M3_M1.append(element)
if 'L1-N3' in x_sections[str(element)]['lines']:
L1_N3.append(x_sections[str(element)]['lines']['L1-N3']['weight']/x_sections[str(element)]['lines']['L3-M5']['weight'])
ele_L1_N3.append(element)
if 'L3-N5' in x_sections[str(element)]['lines']:
L3_N5.append(x_sections[str(element)]['lines']['L3-N5']['weight']/x_sections[str(element)]['lines']['L3-M5']['weight'])
ele_L3_N5.append(element)
if 'L2-M4' in x_sections[str(element)]['lines']:
L2_M4.append(x_sections[str(element)]['lines']['L2-M4']['weight']/x_sections[str(element)]['lines']['L3-M5']['weight'])
ele_L2_M4.append(element)
ele = np.linspace(1,94,94)
plt.figure()
plt.plot(ele_K_M5 ,K_M5, label = 'K-M$_3$ / K-L$_3$')
plt.plot(ele_M3_M1,L3_M1, label = 'L$_3$-M$_1$ / L$_3$-M$_5$')
plt.plot(ele_L1_N3,L1_N3, label = 'L$_1$-N$_3$ / L$_3$-M$_5$')
plt.plot(ele_L3_N5,L3_N5, label = 'L$_3$-N$_5$ / L$_3$-M$_5$')
plt.plot(ele_L2_M4,L2_M4, label = 'L$_2$-M$_4$ / L$_3$-M$_5$')
plt.ylabel('')
plt.gca().set_yscale('log')
plt.xlabel('atomic number')
plt.ylabel('relative weights of lines')
plt.legend();
4.13.6. Characteristic X-ray Intensity#
The cross section
where:
For example for a K shell (Powell 1979):
For silion the characteristic energy or binding energy for the K shell:
The relationship between energy of the exciting electron and critical energy is important and is expressed as overvoltage
The overvoltage of the primary electron with energy
We can express the cross section with the overvoltage:
n_K = 2.0
b_K = 0.35
c_K = 1.0
# Silicon
E_c_Si = 1.838
# Iron
E_c_Fe = 6.401
E = np.linspace(0,30,2048)+0.05
U = E/E_c_Si
Q_Si = 6.51*1e-20*n_K*b_K/(U * E_c_Si**2)* np.log(c_K * U)
U = E/E_c_Fe
Q_Fe = 6.51*1e-20*n_K*b_K/(U * E_c_Fe**2)* np.log(c_K * U)
plt.figure()
plt.plot(E,Q_Si, label ='Si')
plt.plot(E,Q_Fe, label ='Fe')
plt.ylim(0.,Q_Si.max()*1.1)
plt.xlabel('excitation energy (keV)')
plt.ylabel('Q (inizations/[e$^-$ (atoms /cm$^2$)])')
plt.legend();
x_sections['25']['M4']
{'filename': 'None',
'excl before': 5,
'excl after': 50,
'onset': 7.26159,
'factor': 0.6666666666666666}
"""
(Casnati's equation) was found to fit cross-section data to
typically better than +-10% over the range 1<=Uk<=20 and 6<=Z<=79."
Note: This result is for K shell. L & M are much less well characterized.
C. Powell indicated in conversation that he believed that Casnati's
expression was the best available for L & M also.
"""
shell_occupancy={'K':2,'L1':2,'L2':2,'L3':4,'M1':2,'M1':2,'M2':2,'M3':4,'M4':4,'M5':6}
import scipy.constants as const
res = 0.0;
beamE = 30
rel = []
Z= []
shell = 'M4'
for element in range(25,56):
ee = x_sections[str(element)][shell]['onset']#getEdgeEnergy();
u = ee/beamE;
#print(u)
if(u > 1.0):
u2 = u * u;
phi = 10.57 * np.exp((-1.736 / u) + (0.317 / u2));
#psi = Math.pow(ee / PhysicalConstants.RydbergEnergy, -0.0318 + (0.3160 / u) + (-0.1135 / u2));
psi = np.power(ee / const.Rydberg, -0.0318 + (0.3160 / u) + (-0.1135 / u2));
i = ee / const.electron_mass# PhysicalConstants.ElectronRestMass;
t = beamE / const.electron_mass#PhysicalConstants.ElectronRestMass;
f = ((2.0 + i) / (2.0 + t)) * np.sqrt((1.0 + t) / (1.0 + i)) * np.power(((i + t) * (2.0 + t) * np.sqrt(1.0 + i))/ ((t * (2.0 + t) * np.sqrt(1.0 + i)) + (i * (2.0 + i))), 1.5);
getGroundStateOccupancy = shell_occupancy[shell]
res = ((getGroundStateOccupancy * np.sqrt((const.value('Bohr radius') * const.Rydberg) / ee) * f * psi * phi * np.log(u)) / u);
rel.append(res)
Z.append(element)
#print(res)
plt.figure()
plt.title(f"Casnati's cross section for beam energy {beamE:.0f} keV")
plt.plot(Z,rel)
plt.xlabel('atomic number Z')
plt.ylabel('Q [barns]');
Even though the cross section raises fast with overvoltage
We can also see that the intial overvoltage
4.13.7. X-ray Production in a Thick Solid Sample#
If the sample is sufficient thick so that all electrons interact ( several micrometers) the X-ray intensity is found to follow an expression fo the form:
where:
The exponent
n = 1.5
U_0 = np.linspace(1,10, 2048)
i_p = 1.0 # in nA
I = i_p * (U_0-1.0)**n
plt.figure()
plt.plot(U_0,I)
plt.axhline(y=0., color='gray', linewidth=0.5)
plt.xlabel('overvoltage $U_0$ [kV]')
plt.ylabel('$I$ [nA]');
4.13.8. X-ray Production in a Thin Foil#
Now we have all the ingeedients to look at the total generation of X-rays.
The number of photons
The number of X-rays increase linearly with the number of atoms per unit area (
In the graph above, we can see the importance of the overvoltage
4.13.9. Quiz#
Why is the overvoltage increasing so much while the cross section remains rather constant above the threshold?

